Tutorials & Step-by-Step instructions
Step-by-step tutorials on how to run BIGapp, load data, set parameters and more.
BIGapp provides a powerful and intuitive interface for researchers and breeders to analyze genomic data without requiring command-line expertise.
*Please note that at this time files used for BIGapp must be processed by BI. Staff are working on making changes that allows for files that are not processed by BI. We will update as soon as those changes are made. Please feel free to contact us with any questions.

Step-by-step tutorials on how to run BIGapp, load data, set parameters and more.
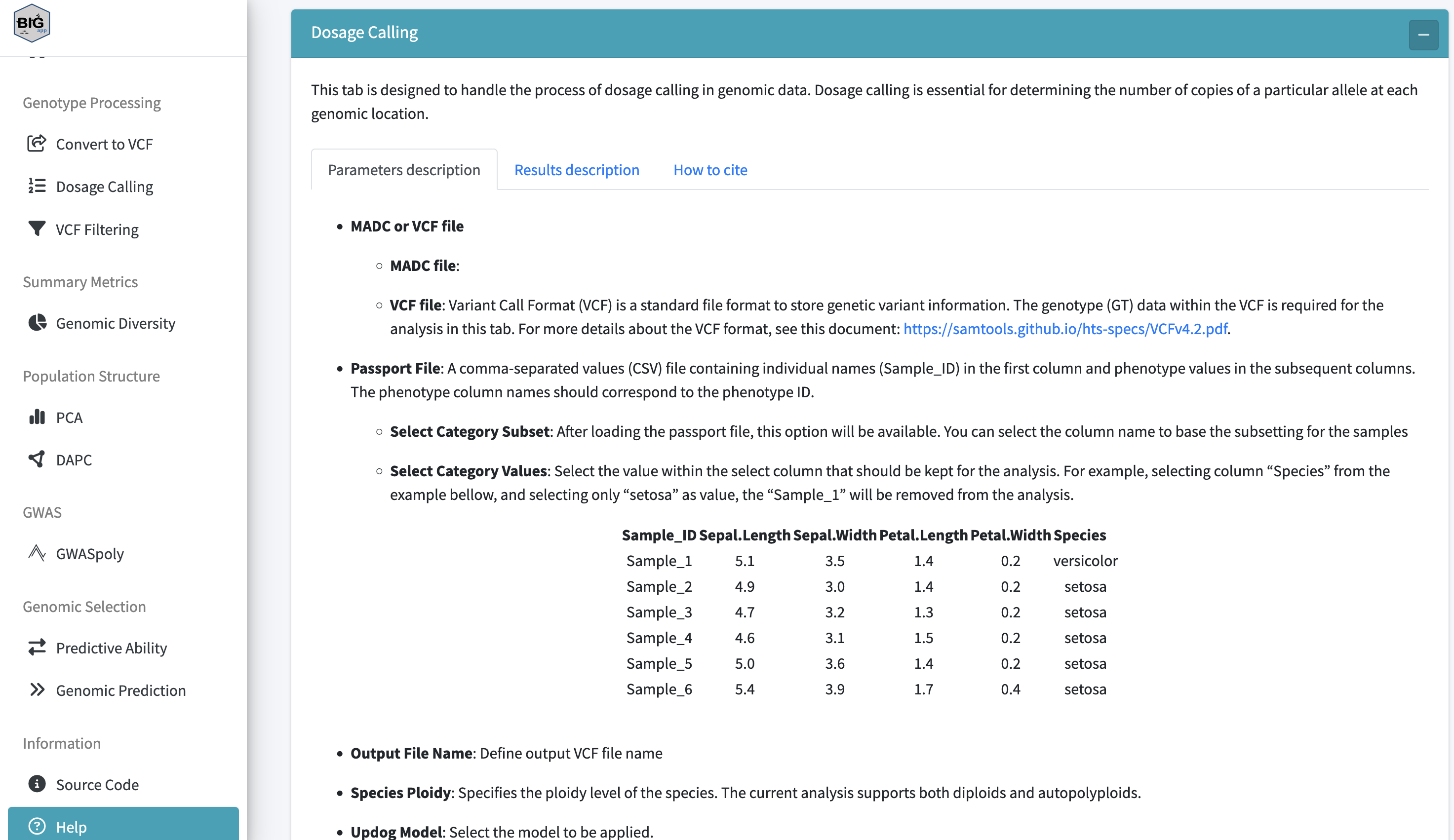
BIGapp features an extensive in-app help section that walks users through the various tools and analysis available in BIGapp.
View the BIGapp code repository and learn more about the technical backend of BIGapp.
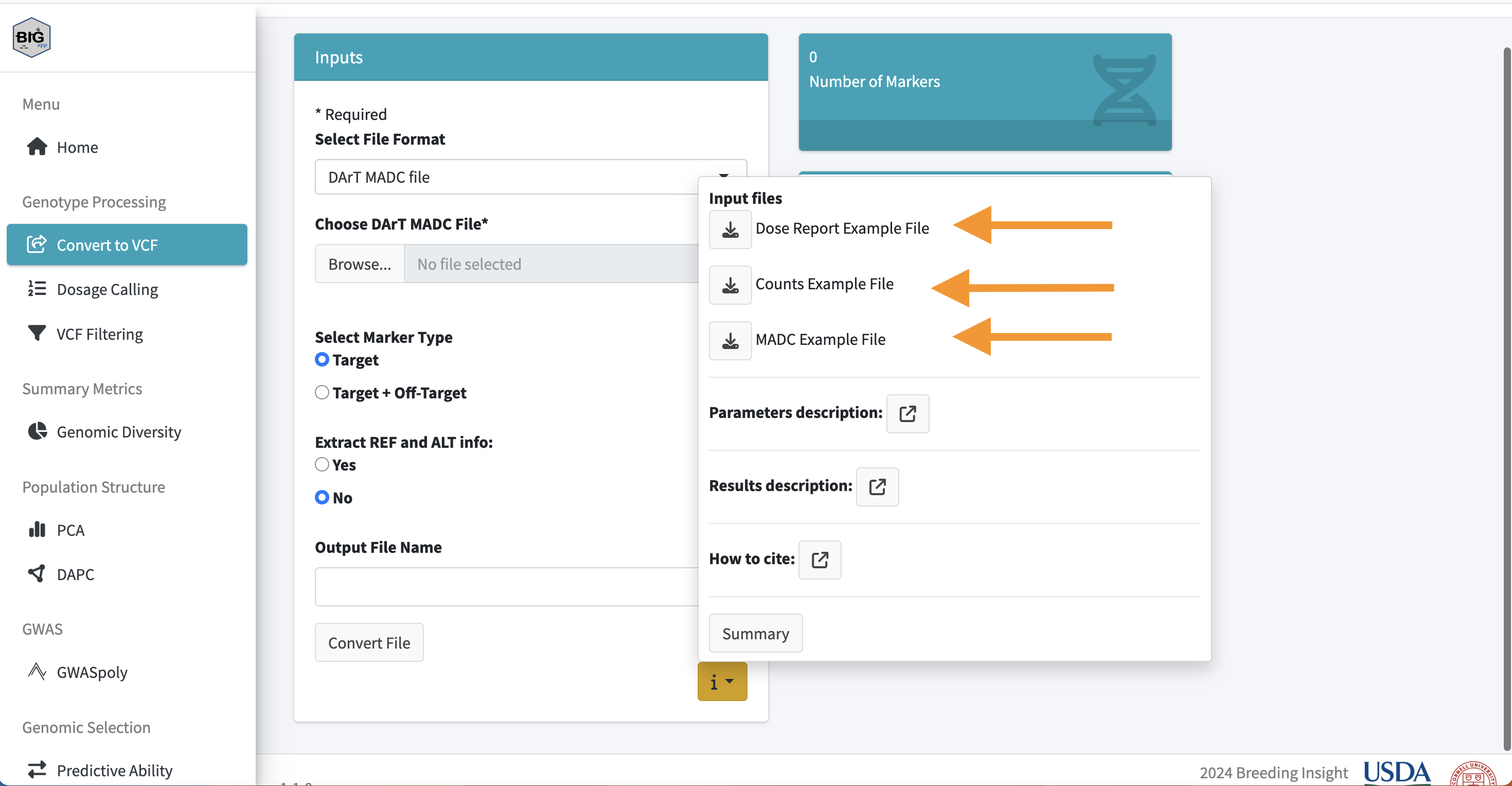
This web demo allows users to move through all the included features of BIGapp to learn more before they download the code. The demo version works best with either the included demo files (see picture below) or less than 500 marker test datasets.
To use the full version of BIGapp, follow the step-by-step tutorials.