About #
Experiments describe the layout of germplasm in space and time, providing the data structure to associate phenotypic observations and genetic markers to germplasm positioned in a field, greenhouse, aquaculture room, growth chamber, 96-well plate, etc. Properly designed, replicated, and randomized experiments can be analyzed and interpreted statistically.
Observation Dataset #
All experiments in DeltaBreed have a top-level dataset, generically referred to as the “observation dataset.” In a randomized experiment this dataset describes the entity into which germplasm are randomly assigned. The top-level experimental entity, the experimental unit, might be represented by a single individual, such as trees randomized within an orchard, or contain multiple individuals, such as within field plots. The top-level experimental dataset is suitable for recording top-level observations and summary statistics derived from repeated measures – time series and/or sub-entity datasets.
ObsUnitIDs #
ObsUnitIDs are system assigned universally unique identifiers (UUIDs) for experimental entities. Top-level experimental entities, or experimental units, must be labeled to facilitate data collection. The ObsUnitID is the appropriate field to use as a barcode to support scan-to-match functions.
Environments & Germplasm #
All experiments are associated with a germplasm list and at least one environment. Environments are defined by their start date and location. Breeders of long-lived species, such as trees, might maintain a single orchard environment for many years. Breeders of short-lived species or animal species with different management requirements at different life stages will have experiments with more discreet environments. For example, as salmon age they are moved through a set of different rooms (environments) containing tanks suited to their size and salinity requirements.
Multi-Environment Experiments
Multi-environment experiments (aka trials) provide information about the adaptability of germplasm to specific environments or to sets of environments. In multi-environmental experiments a set of genotypes or families are raised in a number of environments. Analysis of multi-environmental experiments data can reveal not only which genotypes are ‘best’ overall or in subsets of environments, but also can reveal the relationships among environments in terms of the genotype by environment (GxE) interactions (Isik et al.,2017)
Users can add new environments to a DeltaBreed experiment at any time. Such as to:
- Modify the list of germplasm under evaluation in an experiment, such as when dead plants are replaced with live.
- Create environments to subset observations on long-lived organisms.
Keep in mind that unless an experiment was planned from the start for multi-environment analysis, interpretation of results might be confounded by differences in germplasm age-class between environments.
View #
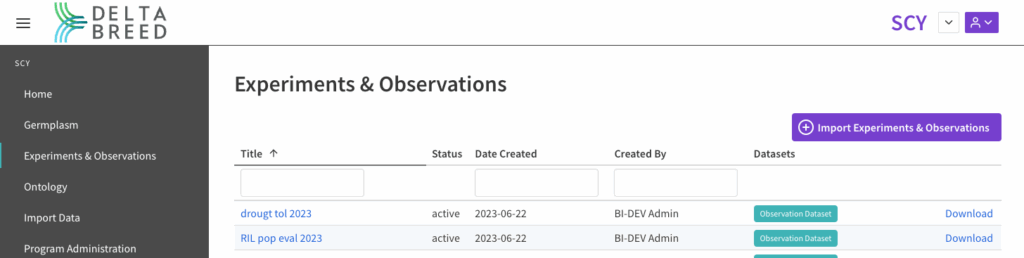
![]() Access experiments and observations from the main left hand menu. Select the name to view the experiment, and “download” to retrieve the data in file format.
Access experiments and observations from the main left hand menu. Select the name to view the experiment, and “download” to retrieve the data in file format.
Download #
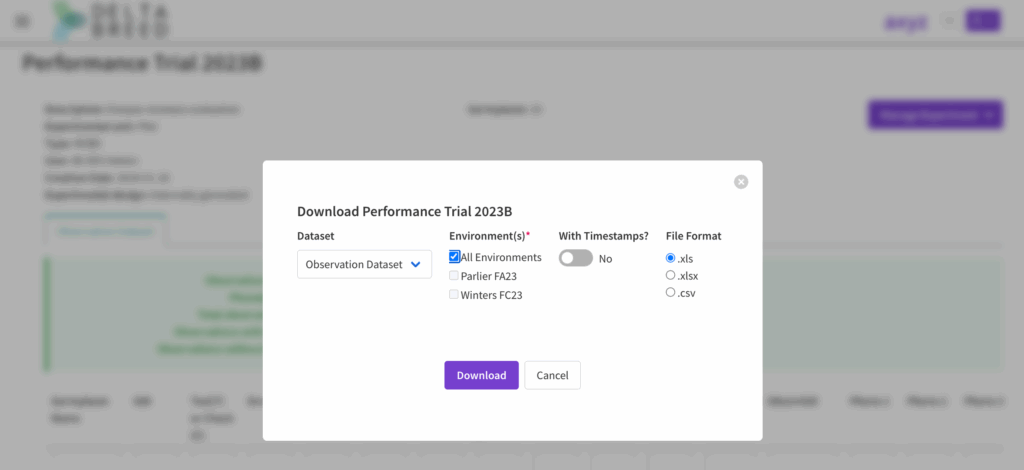
![]() Select download from the Manage Experiment menu. Choose the download options. If you have a multi-environment experiment you can download a single file of all environments or select individual environments. Selecting multiple individual environments will result in the download of a .zip file containing the individual environment files.
Select download from the Manage Experiment menu. Choose the download options. If you have a multi-environment experiment you can download a single file of all environments or select individual environments. Selecting multiple individual environments will result in the download of a .zip file containing the individual environment files.
![]() Open the download file with a program like Excel.
Open the download file with a program like Excel.
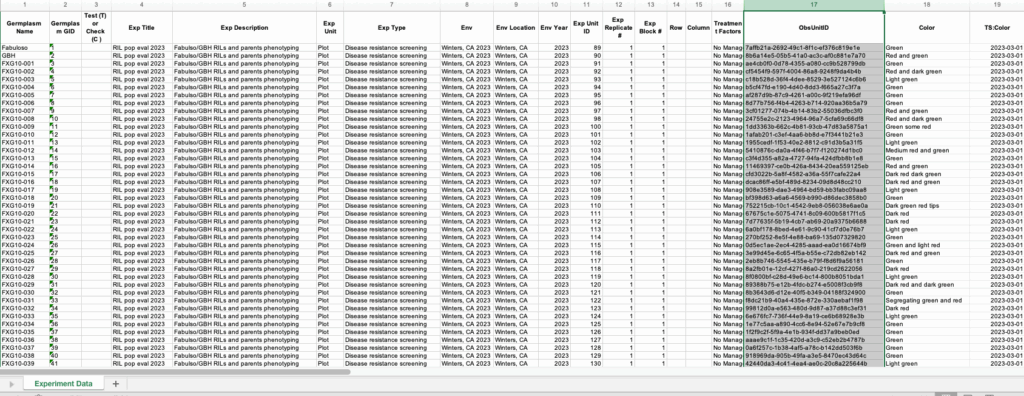
Notice the download file includes the system assigned ObsUnitIDs appropriate for bar coding the experiment unit, in this case plots. Timestamps were included in this export file, denoted by columns with the “TS:” prefix.
Import #
The experimental template can be used to import the experimental design with or without phenotypes and observations.
Experimental germplasm and ontology terms must exist in the system before associated experiments can be created. Therefore, you must import germplasm of interest and create ontology terms before importing an experimental design. GIDs are assigned by the system when a germplasm list is imported and are required in the experimental template. Phenotype names in the experimental import file must perfectly match the name in the DeltaBreed ontology.
Experiment and Observations Template #
![]() From the Import Data menu, open the Experiments & Observations tab. Download the import template (.xls).
From the Import Data menu, open the Experiments & Observations tab. Download the import template (.xls).
![]() Open the .xls template in a program like Excel. The template README defines details of required and optional template data.
Open the .xls template in a program like Excel. The template README defines details of required and optional template data.
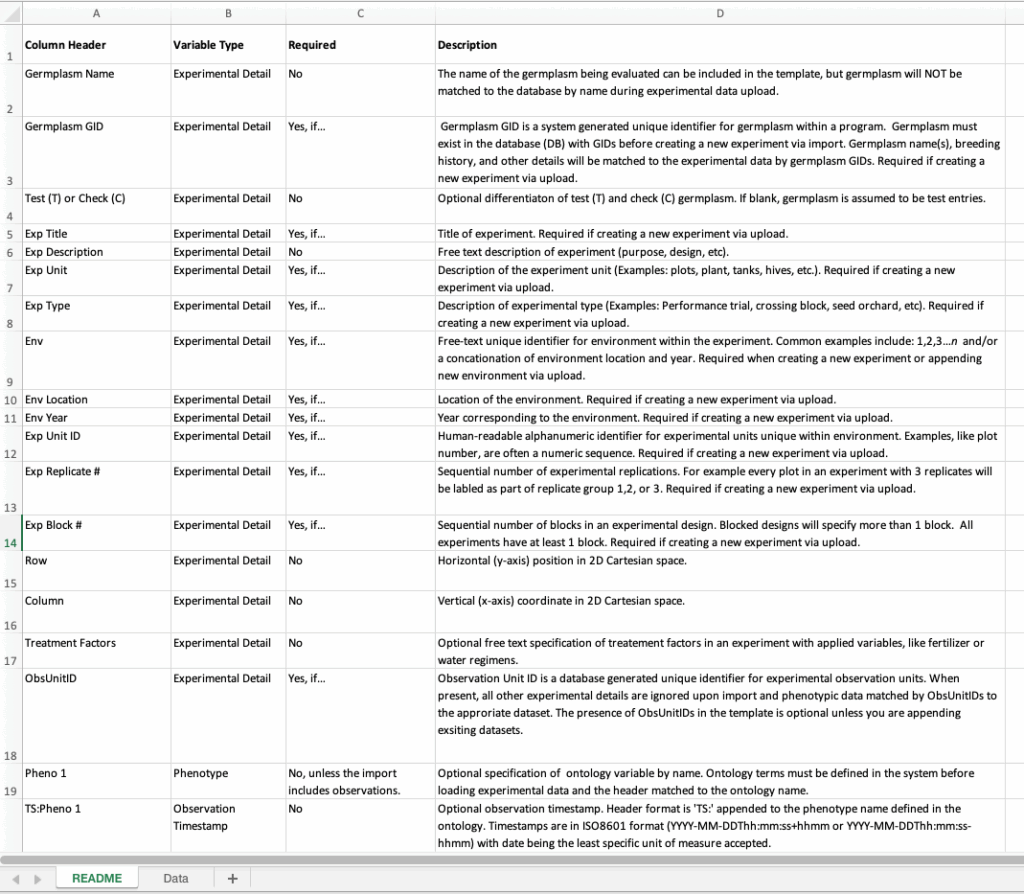
Columns A-W headers define the experimental design – the minimum information needed to create an experiment. The following column headers (W+) allow for specification of phenotypes pre-defined in the ontology.
![]() Edit the “Data” worksheet. System assigned GIDs are required.
Edit the “Data” worksheet. System assigned GIDs are required.
Experimental germplasm MUST exist in the system before associated experiments can be created. Column headings MUST perfectly match the template and the names of ontology terms pre-defined in DeltaBreed. DeltaBreed will give error codes and prevent import if these two conditions are not met.
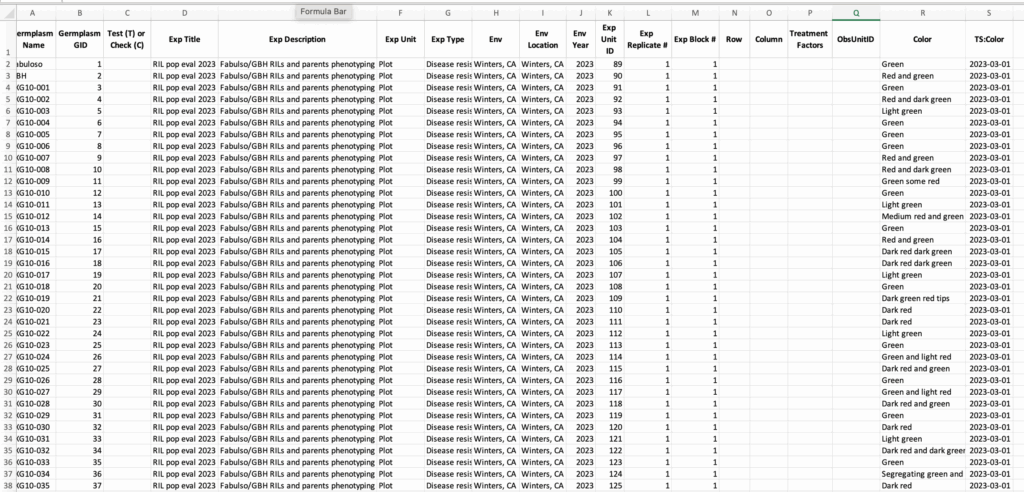
This experimental import contains color phenotyping observations with timestamp denoted by “TS:Color” column.
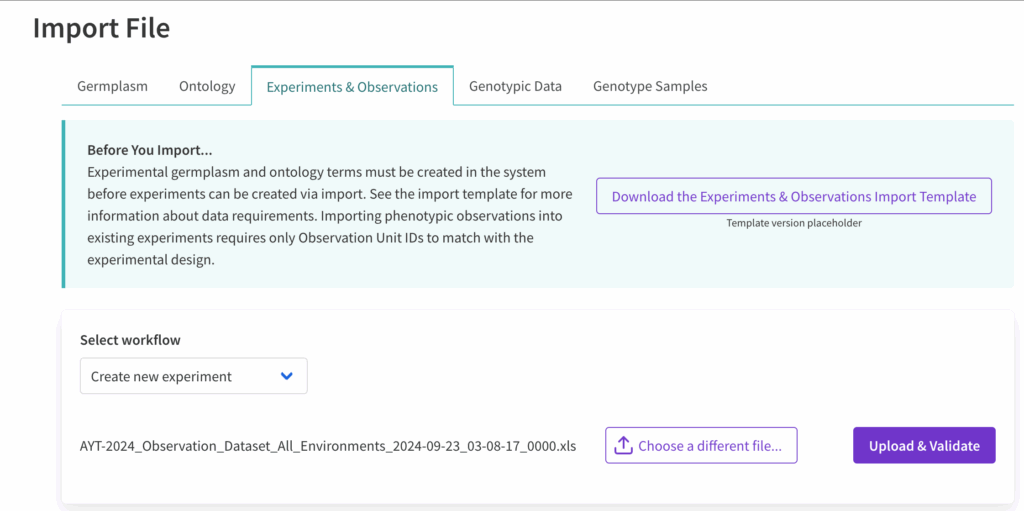
![]() Choose “create new experiment” from the drop down menu. Choose file and import.
Choose “create new experiment” from the drop down menu. Choose file and import.
Observation Validations & Timestamps #
Observations are validated based on the definition of the observation variable as defined in the ontology. For example, if you attempt to import “maybe” for a nominal observation variable defined as “yes” or “no” in the ontology, the system will throw an error. The only exception is “NA” observations, which are allowed for any variable type to denote that an observation could not be made.
The system will accept observations with and without timestamps. Timestamps can be included in the import template by including a column with a heading that appends “TS:” with the observation variable name. For example, observations for the variable Vigor can be timestamped by including a column titled TS:Vigor. Note that if timestamp is included without an observation, it will be ignored and not stored.
Accepted Timestamp Formats #

Preview & Confirm #
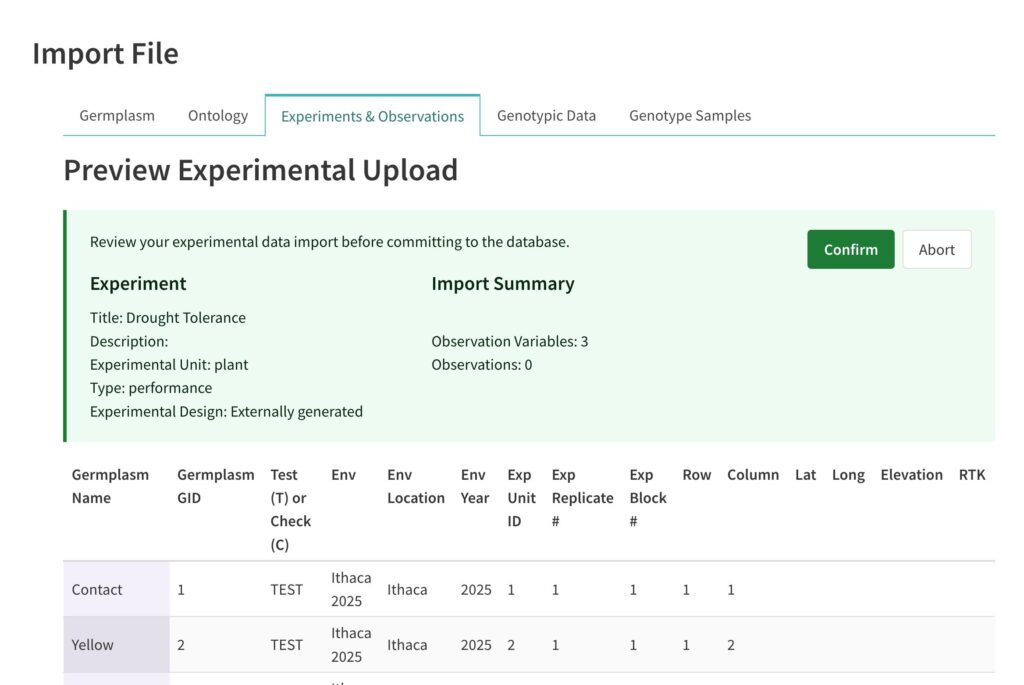
![]() Review the import detail and select Confirm or Abort. Note that committed experimental designs will be assigned ObsUnitIDs by the BI system. Confirmed experiments and their observations are available from Experiments & Observations in the left hand menu.
Review the import detail and select Confirm or Abort. Note that committed experimental designs will be assigned ObsUnitIDs by the BI system. Confirmed experiments and their observations are available from Experiments & Observations in the left hand menu.
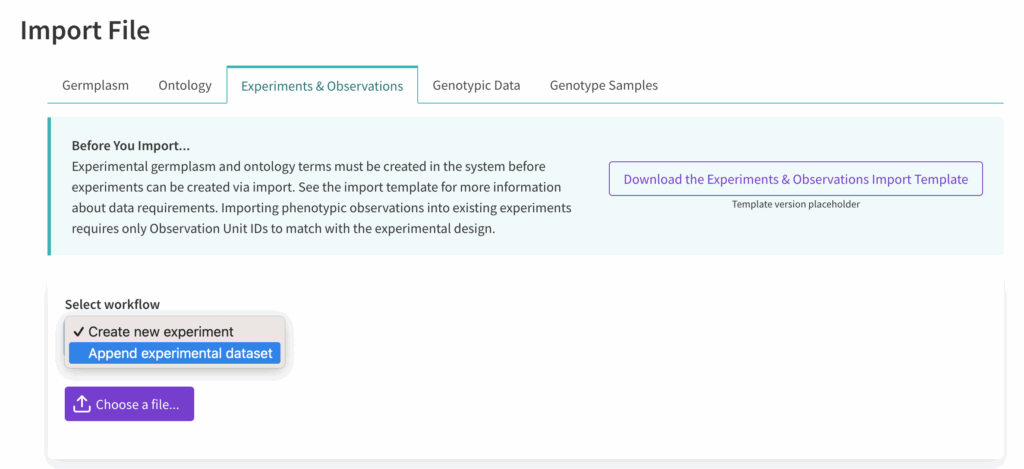
Append Existing Experiment #
Export existing experimental dataset to retrieve a template with system assigned ObsUnitIds. Append the file with new variables and observation.
Overwrite Observations #
- The observation and timestamp can both be overwritten.
- The system keeps a record on the backend of overwritten observations. Contact your system administrator (BI coordinator) if you need to recover erroneously overwritten observations.
- Observations and/or time timestamps proposed for overwriting are validated according to the ontology and accepted timestamp formats.
- Blank cells in the import file will NOT overwrite committed observations.
NAobservations can be used to overwrite any committed observation (case insensitive).
![]() Download the existing experiment from DeltaBreed. The downloaded file will contain ObsUnitIDs and the committed observations.
Download the existing experiment from DeltaBreed. The downloaded file will contain ObsUnitIDs and the committed observations.
![]() Make edits to the observation(s) in the downloaded file with ObsUnitID and save. In the experiments & observations screen, select “append experimental dataset” from the drop down menu.
Make edits to the observation(s) in the downloaded file with ObsUnitID and save. In the experiments & observations screen, select “append experimental dataset” from the drop down menu.
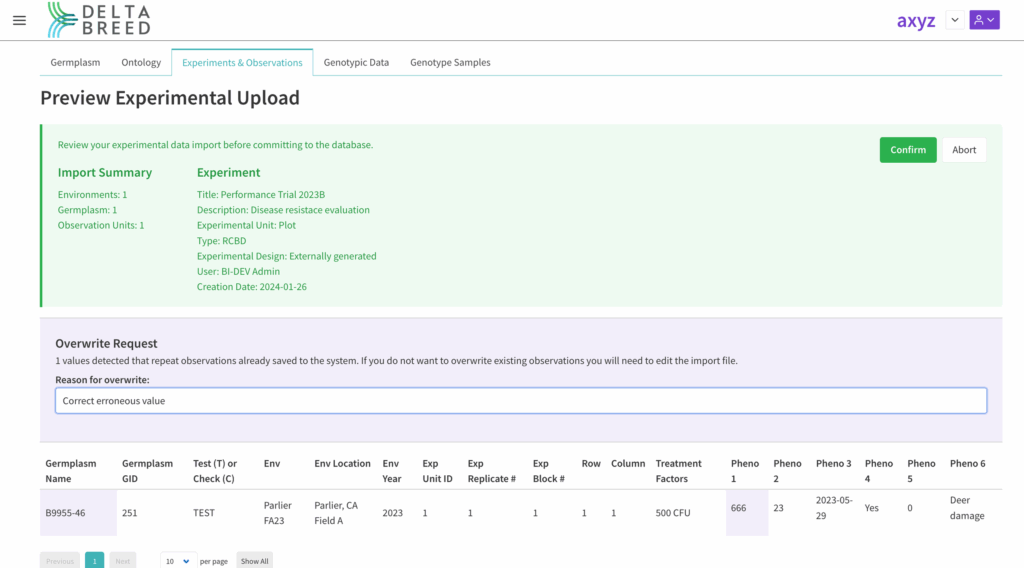
Upon import, the system alerts users if observation(s) are pending overwrite. Enter a reason for the overwrite and select confirm.
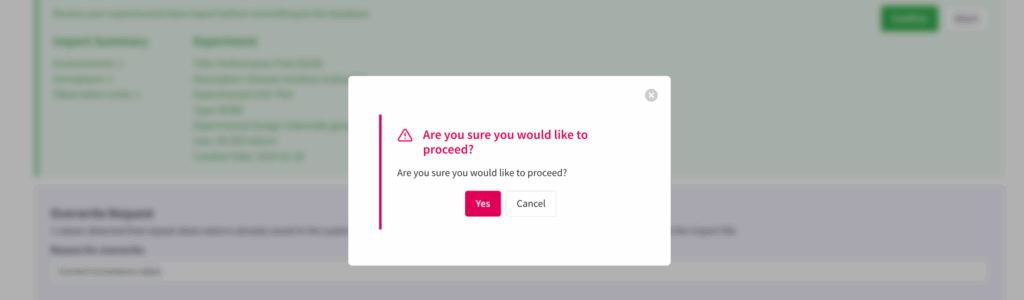
![]() Select Yes if you want to proceed and all of the observations, including any overwrites, will be saved to the system.
Select Yes if you want to proceed and all of the observations, including any overwrites, will be saved to the system.
Append Experiment with New Environment(s) #
New environment(s) can be added to an existing experiment if the import file:
- References the existing experiment title exactly.
- The environment name(s) are new to the experiment.
- Variables and observations can not be created when the new environment is created, these can be appended in a subsequent upload.
Delete #
Only a user with that is a program administrator can delete experiments.
![]() Select Delete Experiment from the Manage Experiment button.
Select Delete Experiment from the Manage Experiment button.
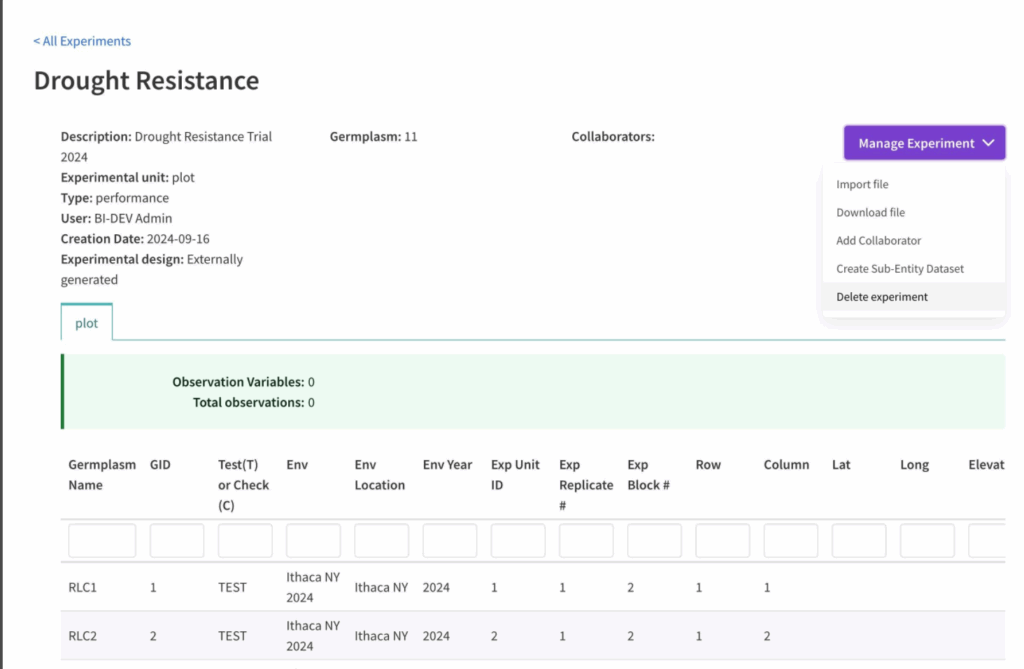
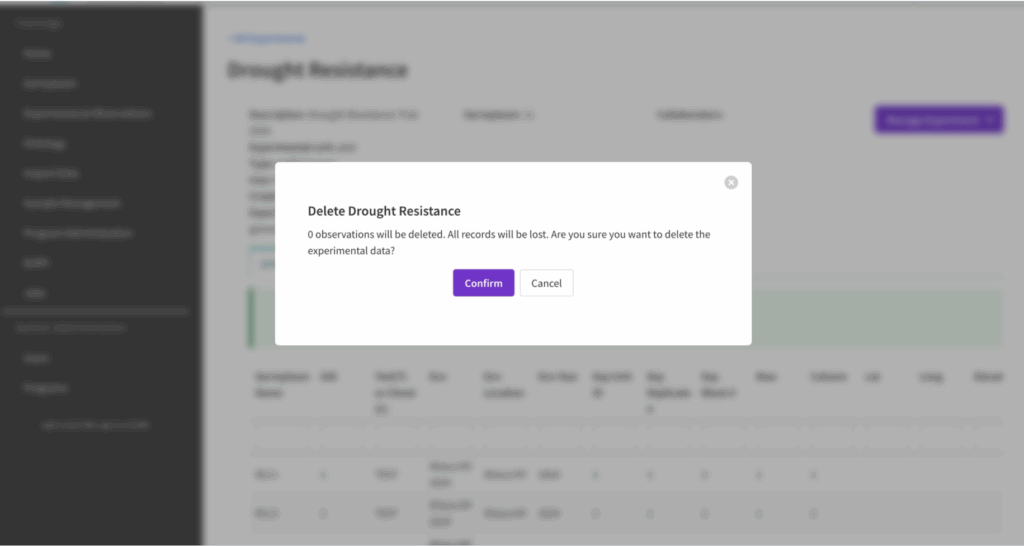
![]() Select Confirm to delete the experiment or Cancel to abort.
Select Confirm to delete the experiment or Cancel to abort.