Genotypic data loading is a working prototype suitable for testing in sandbox programs. Expect to see changes and improvements as we work to integrate our genomic data QC pipeline and haplotype database.
Do not load genotypic data to production programs. Expect genotypic data to be corrupted by future software updates so hold off loading genotypic data.
About #
SNPs identified by genotyping can be visualized in DeltaBreed’s germplasm view. This feature is designed to display marker data coming from Breeding Insight’s QC and validation pipeline. This beta functionality is proof-of-concept that DeltaBreed can load and retrieve genotypic data. Contact your BI coordinator for more information about loading your genotypic data. Expect expansion of genotyping features in future versions of DeltaBreed.
Germplasm View #
![]() Select a genotyped germplasm record and navigate to the details to review the SNP calls. By default the most recently uploaded callset is selected. Previous callsets are available from the dropdown menu. Select views from the different chromosomes, or scaffolds, detailed in the .vcf file.
Select a genotyped germplasm record and navigate to the details to review the SNP calls. By default the most recently uploaded callset is selected. Previous callsets are available from the dropdown menu. Select views from the different chromosomes, or scaffolds, detailed in the .vcf file.
Targeted SNP View #
DeltaBreed can visualize target SNPs, highlighting the differences between the reference genome at the target SNP position and alternative SNPs at the single target base.
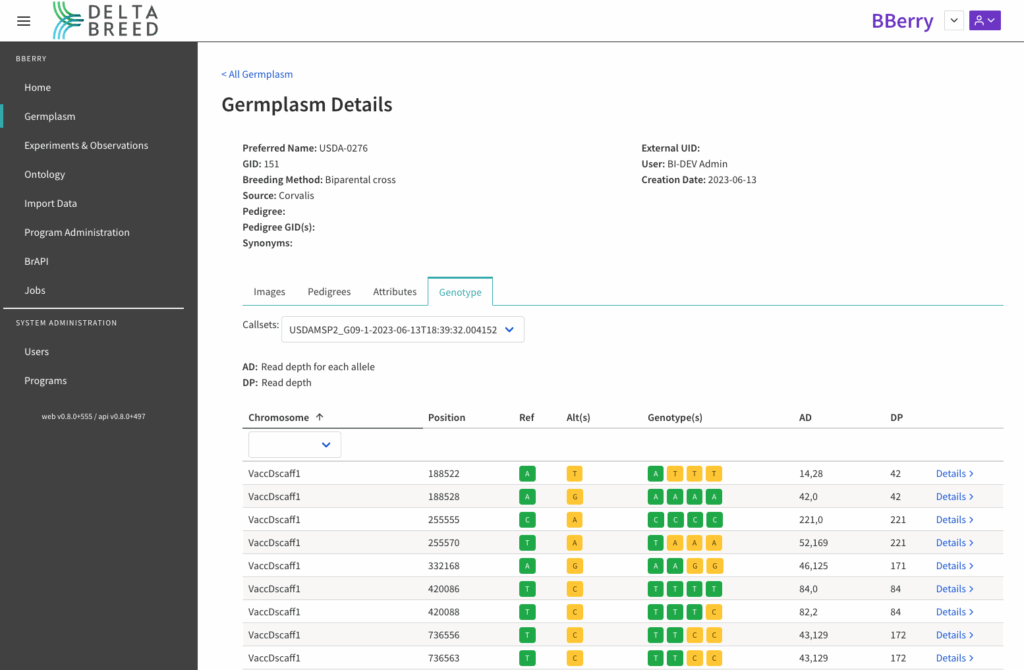
Genotyped tetraploid (n=4) blueberry, chromosome or scaffold view of target SNP calls by position in genome assembly.
Mircohaplotype View #
DeltaBreed can visualize microhaplotypes, short sequence reads around the target SNP. The alternative microhaplotypes represent the diversity identified within your sample submission.
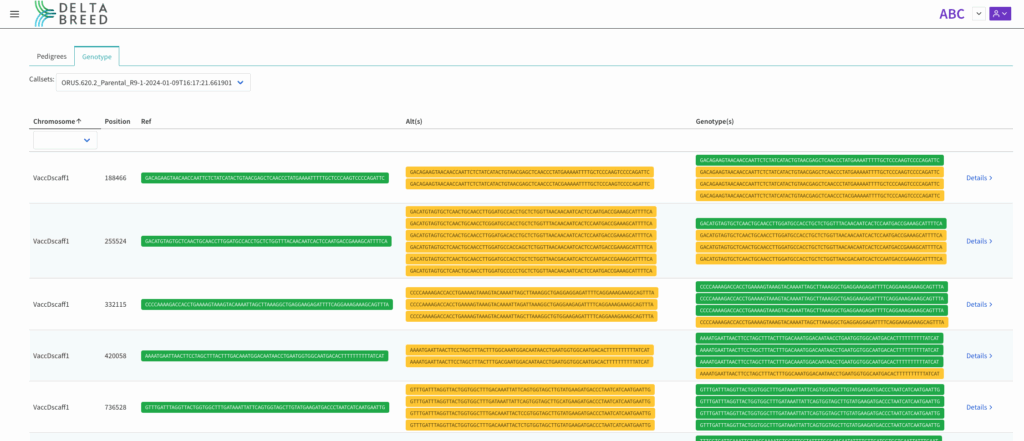
Genotyped tetraploid (n=4) blueberry, chromosome or scaffold view of microhaplotype calls by position in genome assembly.
Create Genotyping Experiment & Import Genotypic Data #
Connecting genotypic to a genotyping experiment is a temporary workflow. In the future expect genotypic data to be connected to a genotyping sample submission.
![]() Edit the experimental template with genotype sample information and import the genotype experiment. (See Experiments & Observations for more information.) Note that the Exp Unit ID should match the sample ID in the VCF file.
Edit the experimental template with genotype sample information and import the genotype experiment. (See Experiments & Observations for more information.) Note that the Exp Unit ID should match the sample ID in the VCF file.
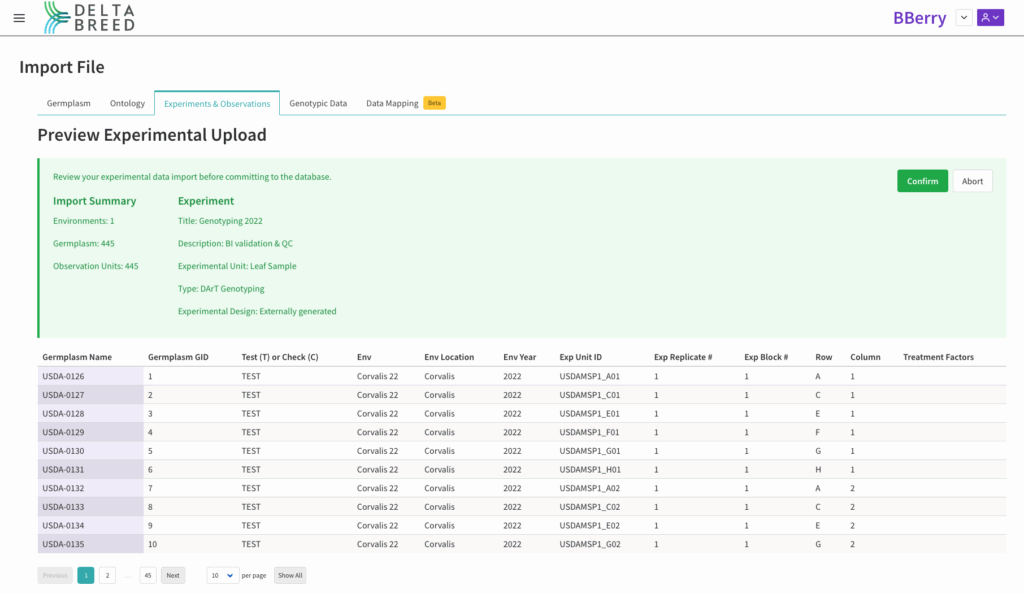
Blueberry genotyping experiment: 445 leaf samples were collected into five 96 well plates. In this example, Exp Block # = plate # and Row and Column correspond to well position.
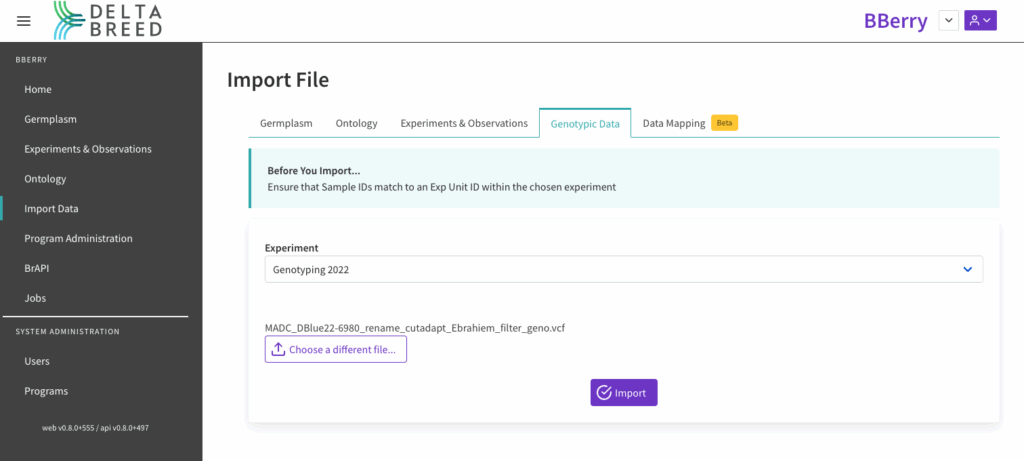
![]() Genotyping data processed through Breeding Insight’s QC and validation pipeline in .vcf format can be loaded via the Import Data module. Select the Genotypic Data experiment and import. The Exp Unit IDs should match the sample ID in the .vcf file.
Genotyping data processed through Breeding Insight’s QC and validation pipeline in .vcf format can be loaded via the Import Data module. Select the Genotypic Data experiment and import. The Exp Unit IDs should match the sample ID in the .vcf file.